Gene-Swaps Could Let Influenza Jump Species
Influenza viruses like bird flu can mix and match their genomes, and this has played a role in at least three of the last four flu pandemics
Avian influenza, a virus of the orthomyxoviridae family. The flu virus causes an infectious and contagious respiratory disease, and often results in a pandemic and/or smaller seasonal epidemic.
James Cavallini/Science Source
Influenza viruses are shifty entities. They accumulate small genetic changes on a regular basis, necessitating yearly updates to the flu vaccines because the prior year’s strain may not look much like the following year’s. But they can also make sudden leaps by incurring big genetic changes that may allow them to jump from one animal species to another or to humans.
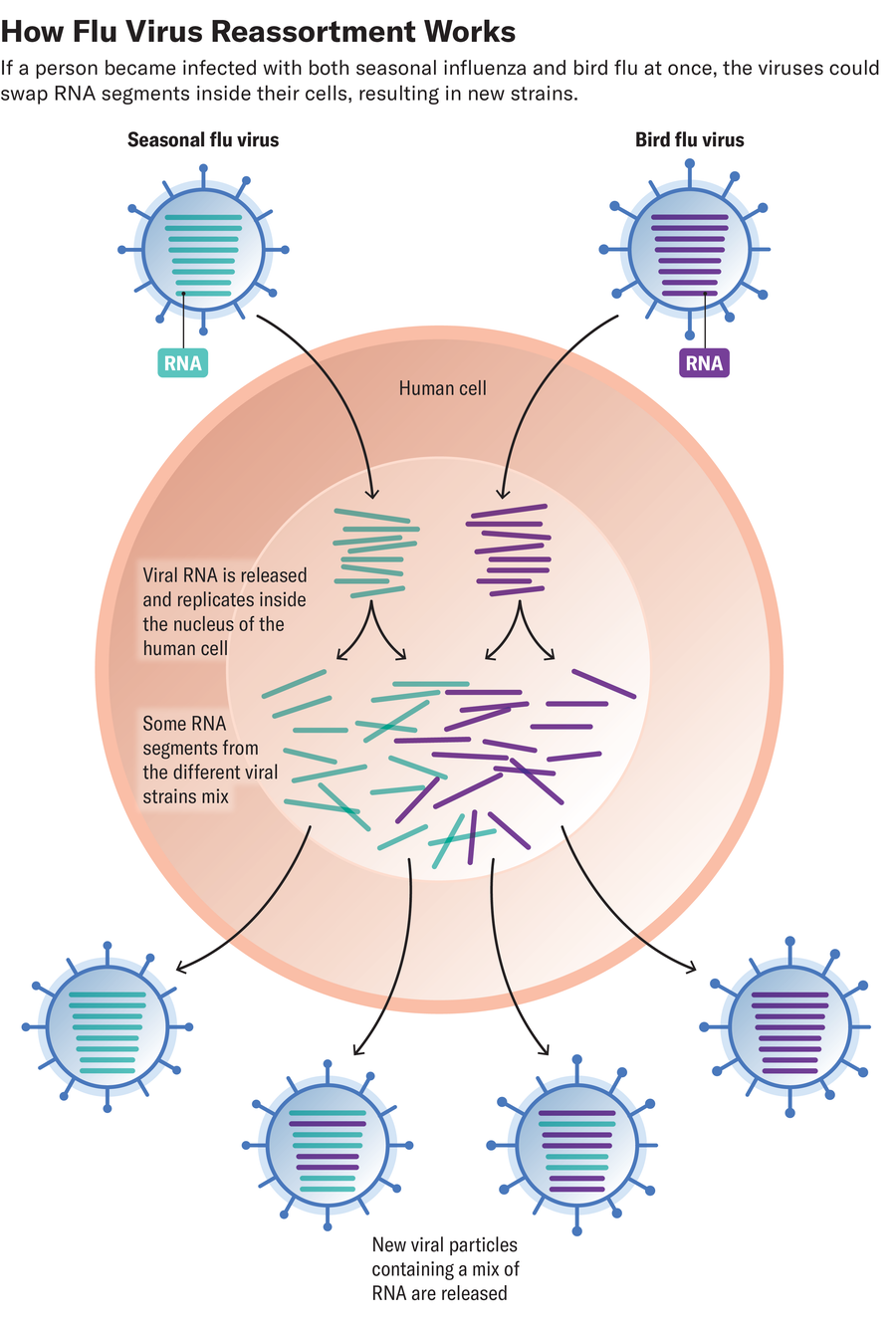
A seemingly ingenious and sneaky way for viruses to make these leaps is by swapping genetic material with other flu strains. Called reassortment, this exchange happens when a person or animal is infected with two types of flu virus at the same time. While replicating inside the host cell, the viruses can grab bits of each other’s genetic code and incorporate them into their own gene sequences.
Reassortment is much less common than small mutations that change the flu year to year, but it’s important: at least three of the last four human flu pandemics have involved reassortment.
On supporting science journalism
If you’re enjoying this article, consider supporting our award-winning journalism by subscribing. By purchasing a subscription you are helping to ensure the future of impactful stories about the discoveries and ideas shaping our world today.
“Reassortment has played a major, major role in the emergence of pandemic influenza,” says Daniel Perez, a professor of poultry medicine at the University of Georgia College of Veterinary Medicine, who studies how flu moves between species.
The past century saw four flu pandemics. The first was the notorious 1918 Great Influenza, which killed around 50 million people. The second was in 1957, when a new flu killed between one million and four million people worldwide. In 1968 another new flu emerged, killing another one million to four million people. Finally, in 2009, a novel swine flu appeared, killing between 151,000 and 575,000 people that year.
Flu viruses are categorized by two types of proteins on their surfaces, hemagglutinin (HA) and neuraminidase (NA). These proteins each have multiple subtypes, which is why you’ll see labels such as H1N1 or H5N1. The H refers to the HA protein type, and the N refers to the type of NA protein. The Great Influenza that swept the globe during World War I was an H1N1 flu that likely emerged in Kansas. Its descendants circulated in both humans and pigs until 1957, when it was suddenly replaced in humans by an H2N2 flu. This new virus first popped up in southern China. Its main genetic backbone belonged to the 1918 flu, Perez says, but it had acquired three new gene sequences from an avian flu, swapping its HA and NA proteins for new subtypes. For reasons not completely understood, this new H2N2 wiped out H1N1 in humans for decades—H1N1 wouldn’t be seen again in people until 1977.
The 1968 pandemic was another reassortment event. This time, the H2N2 that was circulating in humans swapped genes with an H3N2 avian influenza, probably somewhere in China. (The first identified outbreak was in Hong Kong.)
Then came the 2009 pandemic, a true “globalized pandemic,” Perez says. In the early 2000s there had been a few sporadic human infections in the U.S. with so-called triple-reassorted flu viruses that contained genes from human, avian and swine influenzas. These cases were rare and mostly in people who worked on pig farms; these viruses didn’t transmit from human to human. That changed in 2009 when the triple-reassorted viruses picked up new genes from a Eurasian swine flu. “It’s a perfect example of globalization,” Perez says, “because the virus contains not only gene segments from an avian flu, from a swine flu [and] from a human flu but also from very different geographical locations.”
The reassortment of flu viruses that infect different species fortunately happens relatively infrequently, says Charlotte Kristensen, a postdoctoral researcher in veterinary clinical microbiology at the University of Copenhagen. “It has to be two different viruses infecting the same host cell, and the reassortment has to be successful. And it’s not always like the gene segments are compatible,” she says.
Such reassortment happens all the time between avian flu strains that infect birds, says Yuan Liang, also a University of Copenhagen veterinary clinical microbiology postdoctoral researcher. “Especially since 2020, there have been a lot of new variants emerging because of reassortments” in birds, Liang says.
The various strains of H5N1 circulating now in wild birds, domestic poultry and dairy cows have yet to cause a pandemic in people. It’s hard to say whether the virus will stay mostly in animals or whether we’re now in a period like the one before the 2009 flu pandemic, when farmworkers occasionally came down with a reassorted virus that would later gain the gene sequences it needed to spread from person to person. No one expected H5N1 to take hold in dairy cattle, Liang says, so the question now is what new, unexpected step this virus might take.
“This whole situation really highlights how little we know and how complex it is,” Kristensen says.